Probabilistic Modelling using the Infinite Mixture Model
In many applications it is desirable to allow the model to adjust its complexity to the amount of data. Consider for example the task of assigning objects into clusters or groups. This task often involves the specification of the number of groups. However, often times it is not known beforehand how many groups exist. Moreover, in some applictions, e.g. modelling topics in text documents or grouping species, the number of examples per group is heavy tailed. This makes it impossible to predefine the number of groups and requiring the model to form new groups when data points from previously unseen groups are observed.
A natural approach for such applications is the use of non-parametric models. This tutorial will introduce how to use the Dirichlet process in a mixture of infinitely many Gaussians using Turing. For further information on Bayesian nonparametrics and the Dirichlet process we refer to the introduction by Zoubin Ghahramani and the book "Fundamentals of Nonparametric Bayesian Inference" by Subhashis Ghosal and Aad van der Vaart.
using Turing
Mixture Model
Before introducing infinite mixture models in Turing, we will briefly review the construction of finite mixture models. Subsequently, we will define how to use the Chinese restaurant process construction of a Dirichlet process for non-parametric clustering.
Two-Component Model
First, consider the simple case of a mixture model with two Gaussian components with fixed covariance. The generative process of such a model can be written as:
\begin{equation*} \begin{aligned} \pi_1 &\sim \mathrm{Beta}(a, b) \ \pi_2 &= 1-\pi_1 \ \mu_1 &\sim \mathrm{Normal}(\mu_0, \Sigma_0) \ \mu_2 &\sim \mathrm{Normal}(\mu_0, \Sigma_0) \ z_i &\sim \mathrm{Categorical}(\pi_1, \pi_2) \ x_i &\sim \mathrm{Normal}(\mu_{z_i}, \Sigma) \end{aligned} \end{equation*}
where $\pi_1, \pi_2$ are the mixing weights of the mixture model, i.e. $\pi_1 + \pi_2 = 1$, and $z_i$ is a latent assignment of the observation $x_i$ to a component (Gaussian).
We can implement this model in Turing for 1D data as follows:
@model function two_model(x)
# Hyper-parameters
μ0 = 0.0
σ0 = 1.0
# Draw weights.
π1 ~ Beta(1, 1)
π2 = 1 - π1
# Draw locations of the components.
μ1 ~ Normal(μ0, σ0)
μ2 ~ Normal(μ0, σ0)
# Draw latent assignment.
z ~ Categorical([π1, π2])
# Draw observation from selected component.
if z == 1
x ~ Normal(μ1, 1.0)
else
x ~ Normal(μ2, 1.0)
end
end
two_model (generic function with 2 methods)
Finite Mixture Model
If we have more than two components, this model can elegantly be extended using a Dirichlet distribution as prior for the mixing weights $\pi_1, \dots, \pi_K$. Note that the Dirichlet distribution is the multivariate generalization of the beta distribution. The resulting model can be written as:
$$ \begin{align} (\pi_1, \dots, \pi_K) &\sim Dirichlet(K, \alpha) \ \mu_k &\sim \mathrm{Normal}(\mu_0, \Sigma_0), ;; \forall k \ z &\sim Categorical(\pi_1, \dots, \pi_K) \ x &\sim \mathrm{Normal}(\mu_z, \Sigma) \end{align} $$
which resembles the model in the Gaussian mixture model tutorial with a slightly different notation.
Infinite Mixture Model
The question now arises, is there a generalization of a Dirichlet distribution for which the dimensionality $K$ is infinite, i.e. $K = \infty$?
But first, to implement an infinite Gaussian mixture model in Turing, we first need to load the Turing.RandomMeasures module. RandomMeasures contains a variety of tools useful in nonparametrics.
using Turing.RandomMeasures
We now will utilize the fact that one can integrate out the mixing weights in a Gaussian mixture model allowing us to arrive at the Chinese restaurant process construction. See Carl E. Rasmussen: The Infinite Gaussian Mixture Model, NIPS (2000) for details.
In fact, if the mixing weights are integrated out, the conditional prior for the latent variable $z$ is given by:
$$ p(z_i = k \mid z_{\not i}, \alpha) = \frac{n_k + \alpha K}{N - 1 + \alpha} $$
where $z_{\not i}$ are the latent assignments of all observations except observation $i$. Note that we use $n_k$ to denote the number of observations at component $k$ excluding observation $i$. The parameter $\alpha$ is the concentration parameter of the Dirichlet distribution used as prior over the mixing weights.
Chinese Restaurant Process
To obtain the Chinese restaurant process construction, we can now derive the conditional prior if $K \rightarrow \infty$.
For $n_k > 0$ we obtain:
$$ p(z_i = k \mid z_{\not i}, \alpha) = \frac{n_k}{N - 1 + \alpha} $$
and for all infinitely many clusters that are empty (combined) we get:
$$ p(z_i = k \mid z_{\not i}, \alpha) = \frac{\alpha}{N - 1 + \alpha} $$
Those equations show that the conditional prior for component assignments is proportional to the number of such observations, meaning that the Chinese restaurant process has a rich get richer property.
To get a better understanding of this property, we can plot the cluster choosen by for each new observation drawn from the conditional prior.
# Concentration parameter.
α = 10.0
# Random measure, e.g. Dirichlet process.
rpm = DirichletProcess(α)
# Cluster assignments for each observation.
z = Vector{Int}()
# Maximum number of observations we observe.
Nmax = 500
for i in 1:Nmax
# Number of observations per cluster.
K = isempty(z) ? 0 : maximum(z)
nk = Vector{Int}(map(k -> sum(z .== k), 1:K))
# Draw new assignment.
push!(z, rand(ChineseRestaurantProcess(rpm, nk)))
end
using Plots
# Plot the cluster assignments over time
@gif for i in 1:Nmax
scatter(
collect(1:i),
z[1:i];
markersize=2,
xlabel="observation (i)",
ylabel="cluster (k)",
legend=false,
)
end
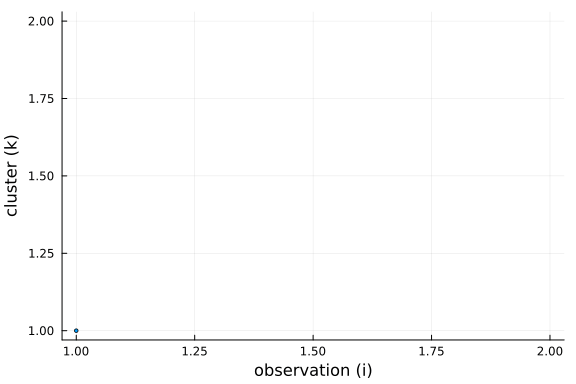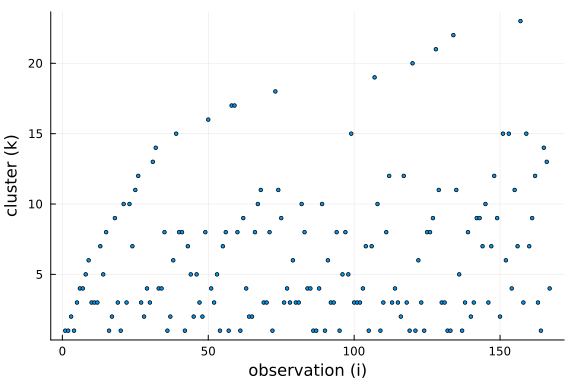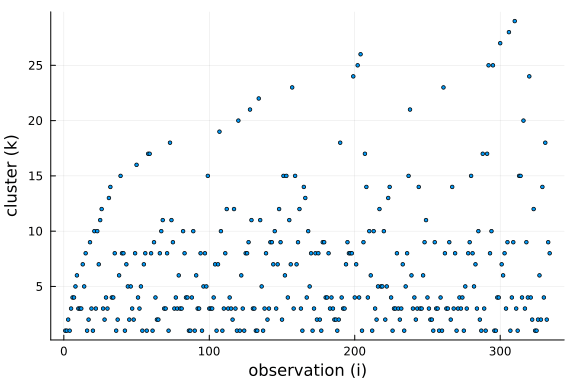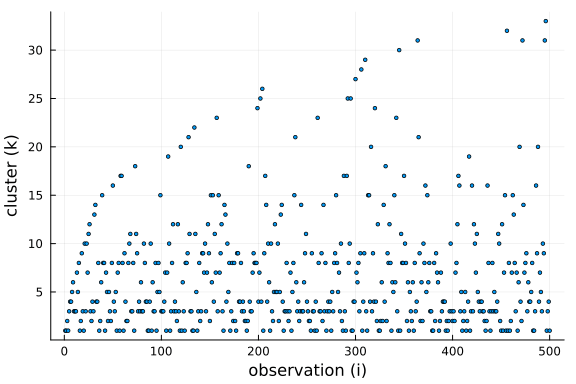
Further, we can see that the number of clusters is logarithmic in the number of observations and data points. This is a side-effect of the "rich-get-richer" phenomenon, i.e. we expect large clusters and thus the number of clusters has to be smaller than the number of observations.
$$ \mathbb{E}[K \mid N] \approx \alpha \cdot log \big(1 + \frac{N}{\alpha}\big) $$
We can see from the equation that the concentration parameter $\alpha$ allows us to control the number of clusters formed a priori.
In Turing we can implement an infinite Gaussian mixture model using the Chinese restaurant process construction of a Dirichlet process as follows:
@model function infiniteGMM(x)
# Hyper-parameters, i.e. concentration parameter and parameters of H.
α = 1.0
μ0 = 0.0
σ0 = 1.0
# Define random measure, e.g. Dirichlet process.
rpm = DirichletProcess(α)
# Define the base distribution, i.e. expected value of the Dirichlet process.
H = Normal(μ0, σ0)
# Latent assignment.
z = tzeros(Int, length(x))
# Locations of the infinitely many clusters.
μ = tzeros(Float64, 0)
for i in 1:length(x)
# Number of clusters.
K = maximum(z)
nk = Vector{Int}(map(k -> sum(z .== k), 1:K))
# Draw the latent assignment.
z[i] ~ ChineseRestaurantProcess(rpm, nk)
# Create a new cluster?
if z[i] > K
push!(μ, 0.0)
# Draw location of new cluster.
μ[z[i]] ~ H
end
# Draw observation.
x[i] ~ Normal(μ[z[i]], 1.0)
end
end
infiniteGMM (generic function with 2 methods)
We can now use Turing to infer the assignments of some data points. First, we will create some random data that comes from three clusters, with means of 0, -5, and 10.
using Plots, Random
# Generate some test data.
Random.seed!(1)
data = vcat(randn(10), randn(10) .- 5, randn(10) .+ 10)
data .-= mean(data)
data /= std(data);
Next, we'll sample from our posterior using SMC.
# MCMC sampling
Random.seed!(2)
iterations = 1000
model_fun = infiniteGMM(data);
chain = sample(model_fun, SMC(), iterations);
Finally, we can plot the number of clusters in each sample.
# Extract the number of clusters for each sample of the Markov chain.
k = map(
t -> length(unique(vec(chain[t, MCMCChains.namesingroup(chain, :z), :].value))),
1:iterations,
);
# Visualize the number of clusters.
plot(k; xlabel="Iteration", ylabel="Number of clusters", label="Chain 1")
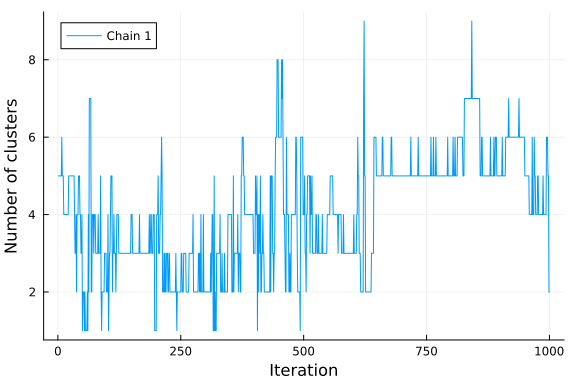
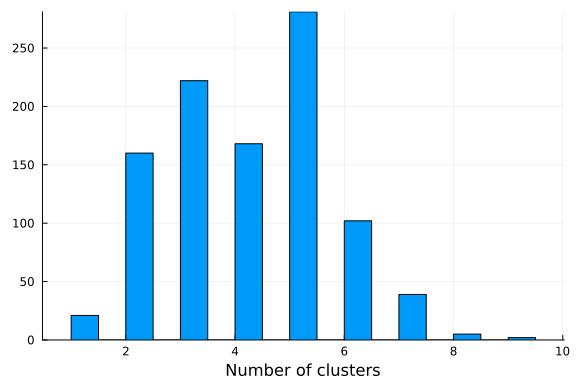
If we visualize the histogram of the number of clusters sampled from our posterior, we observe that the model seems to prefer 3 clusters, which is the true number of clusters. Note that the number of clusters in a Dirichlet process mixture model is not limited a priori and will grow to infinity with probability one. However, if conditioned on data the posterior will concentrate on a finite number of clusters enforcing the resulting model to have a finite amount of clusters. It is, however, not given that the posterior of a Dirichlet process Gaussian mixture model converges to the true number of clusters, given that data comes from a finite mixture model. See Jeffrey Miller and Matthew Harrison: A simple example of Dirichlet process mixture inconsitency for the number of components for details.
histogram(k; xlabel="Number of clusters", legend=false)
One issue with the Chinese restaurant process construction is that the number of latent parameters we need to sample scales with the number of observations. It may be desirable to use alternative constructions in certain cases. Alternative methods of constructing a Dirichlet process can be employed via the following representations:
Size-Biased Sampling Process
$$ j_k \sim \mathrm{Beta}(1, \alpha) \cdot \mathrm{surplus} $$
Stick-Breaking Process $$ v_k \sim \mathrm{Beta}(1, \alpha) $$
Chinese Restaurant Process
$$
p(z_n = k | z_{1:n-1}) \propto \begin{cases}
\frac{m_k}{n-1+\alpha}, \text{ if } m_k > 0\
\frac{\alpha}{n-1+\alpha}
\end{cases}
$$
For more details see this article.
Appendix
These tutorials are a part of the TuringTutorials repository, found at: https://github.com/TuringLang/TuringTutorials.
To locally run this tutorial, do the following commands:
using TuringTutorials
TuringTutorials.weave("06-infinite-mixture-model", "06_infinite-mixture-model.jmd")
Computer Information:
Julia Version 1.9.2
Commit e4ee485e909 (2023-07-05 09:39 UTC)
Platform Info:
OS: Linux (x86_64-linux-gnu)
CPU: 128 × AMD EPYC 7502 32-Core Processor
WORD_SIZE: 64
LIBM: libopenlibm
LLVM: libLLVM-14.0.6 (ORCJIT, znver2)
Threads: 1 on 128 virtual cores
Environment:
JULIA_CPU_THREADS = 128
JULIA_DEPOT_PATH = /cache/julia-buildkite-plugin/depots/7aa0085e-79a4-45f3-a5bd-9743c91cf3da
Package Information:
Status `/cache/build/exclusive-amdci1-0/julialang/turingtutorials/tutorials/06-infinite-mixture-model/Project.toml`
[91a5bcdd] Plots v1.38.17
[fce5fe82] Turing v0.28.1
[9a3f8284] Random
And the full manifest:
Status `/cache/build/exclusive-amdci1-0/julialang/turingtutorials/tutorials/06-infinite-mixture-model/Manifest.toml`
[47edcb42] ADTypes v0.1.6
[621f4979] AbstractFFTs v1.5.0
[80f14c24] AbstractMCMC v4.4.2
⌅ [7a57a42e] AbstractPPL v0.5.4
[1520ce14] AbstractTrees v0.4.4
[79e6a3ab] Adapt v3.6.2
[0bf59076] AdvancedHMC v0.5.3
[5b7e9947] AdvancedMH v0.7.5
[576499cb] AdvancedPS v0.4.3
[b5ca4192] AdvancedVI v0.2.4
[dce04be8] ArgCheck v2.3.0
[4fba245c] ArrayInterface v7.4.11
[a9b6321e] Atomix v0.1.0
[13072b0f] AxisAlgorithms v1.0.1
[39de3d68] AxisArrays v0.4.7
[198e06fe] BangBang v0.3.39
[9718e550] Baselet v0.1.1
⌅ [76274a88] Bijectors v0.12.8
[d1d4a3ce] BitFlags v0.1.7
[fa961155] CEnum v0.4.2
[49dc2e85] Calculus v0.5.1
[082447d4] ChainRules v1.53.0
[d360d2e6] ChainRulesCore v1.16.0
[9e997f8a] ChangesOfVariables v0.1.8
[944b1d66] CodecZlib v0.7.2
[35d6a980] ColorSchemes v3.23.0
[3da002f7] ColorTypes v0.11.4
[c3611d14] ColorVectorSpace v0.10.0
[5ae59095] Colors v0.12.10
[861a8166] Combinatorics v1.0.2
[38540f10] CommonSolve v0.2.4
[bbf7d656] CommonSubexpressions v0.3.0
[34da2185] Compat v4.9.0
[a33af91c] CompositionsBase v0.1.2
[f0e56b4a] ConcurrentUtilities v2.2.1
[88cd18e8] ConsoleProgressMonitor v0.1.2
[187b0558] ConstructionBase v1.5.3
[d38c429a] Contour v0.6.2
[a8cc5b0e] Crayons v4.1.1
[9a962f9c] DataAPI v1.15.0
[864edb3b] DataStructures v0.18.15
[e2d170a0] DataValueInterfaces v1.0.0
[244e2a9f] DefineSingletons v0.1.2
[8bb1440f] DelimitedFiles v1.9.1
[b429d917] DensityInterface v0.4.0
[163ba53b] DiffResults v1.1.0
[b552c78f] DiffRules v1.15.1
[31c24e10] Distributions v0.25.100
[ced4e74d] DistributionsAD v0.6.52
[ffbed154] DocStringExtensions v0.9.3
[fa6b7ba4] DualNumbers v0.6.8
⌃ [366bfd00] DynamicPPL v0.23.0
[cad2338a] EllipticalSliceSampling v1.1.0
[4e289a0a] EnumX v1.0.4
[460bff9d] ExceptionUnwrapping v0.1.9
[e2ba6199] ExprTools v0.1.10
[c87230d0] FFMPEG v0.4.1
[7a1cc6ca] FFTW v1.7.1
[1a297f60] FillArrays v1.5.0
[53c48c17] FixedPointNumbers v0.8.4
[59287772] Formatting v0.4.2
[f6369f11] ForwardDiff v0.10.36
[069b7b12] FunctionWrappers v1.1.3
[77dc65aa] FunctionWrappersWrappers v0.1.3
[d9f16b24] Functors v0.4.5
[46192b85] GPUArraysCore v0.1.5
[28b8d3ca] GR v0.72.9
[42e2da0e] Grisu v1.0.2
[cd3eb016] HTTP v1.9.14
[34004b35] HypergeometricFunctions v0.3.23
[22cec73e] InitialValues v0.3.1
[505f98c9] InplaceOps v0.3.0
[a98d9a8b] Interpolations v0.14.7
[8197267c] IntervalSets v0.7.7
[3587e190] InverseFunctions v0.1.12
[41ab1584] InvertedIndices v1.3.0
[92d709cd] IrrationalConstants v0.2.2
[c8e1da08] IterTools v1.8.0
[82899510] IteratorInterfaceExtensions v1.0.0
[1019f520] JLFzf v0.1.5
[692b3bcd] JLLWrappers v1.4.1
[682c06a0] JSON v0.21.4
[63c18a36] KernelAbstractions v0.9.8
[5ab0869b] KernelDensity v0.6.7
[929cbde3] LLVM v6.1.0
[8ac3fa9e] LRUCache v1.4.1
[b964fa9f] LaTeXStrings v1.3.0
[23fbe1c1] Latexify v0.16.1
[50d2b5c4] Lazy v0.15.1
[1d6d02ad] LeftChildRightSiblingTrees v0.2.0
[6f1fad26] Libtask v0.8.6
[6fdf6af0] LogDensityProblems v2.1.1
[996a588d] LogDensityProblemsAD v1.6.1
[2ab3a3ac] LogExpFunctions v0.3.24
[e6f89c97] LoggingExtras v1.0.0
[c7f686f2] MCMCChains v6.0.3
[be115224] MCMCDiagnosticTools v0.3.5
[e80e1ace] MLJModelInterface v1.8.0
[1914dd2f] MacroTools v0.5.10
[dbb5928d] MappedArrays v0.4.2
[739be429] MbedTLS v1.1.7
[442fdcdd] Measures v0.3.2
[128add7d] MicroCollections v0.1.4
[e1d29d7a] Missings v1.1.0
[872c559c] NNlib v0.9.4
[77ba4419] NaNMath v1.0.2
[86f7a689] NamedArrays v0.9.8
[c020b1a1] NaturalSort v1.0.0
[6fe1bfb0] OffsetArrays v1.12.10
[4d8831e6] OpenSSL v1.4.1
[3bd65402] Optimisers v0.2.19
[bac558e1] OrderedCollections v1.6.2
[90014a1f] PDMats v0.11.17
[69de0a69] Parsers v2.7.2
[b98c9c47] Pipe v1.3.0
[ccf2f8ad] PlotThemes v3.1.0
[995b91a9] PlotUtils v1.3.5
[91a5bcdd] Plots v1.38.17
[aea7be01] PrecompileTools v1.1.2
[21216c6a] Preferences v1.4.0
[08abe8d2] PrettyTables v2.2.7
[33c8b6b6] ProgressLogging v0.1.4
[92933f4c] ProgressMeter v1.7.2
[1fd47b50] QuadGK v2.8.2
[74087812] Random123 v1.6.1
[e6cf234a] RandomNumbers v1.5.3
[b3c3ace0] RangeArrays v0.3.2
[c84ed2f1] Ratios v0.4.5
[c1ae055f] RealDot v0.1.0
[3cdcf5f2] RecipesBase v1.3.4
[01d81517] RecipesPipeline v0.6.12
[731186ca] RecursiveArrayTools v2.38.7
[189a3867] Reexport v1.2.2
[05181044] RelocatableFolders v1.0.0
[ae029012] Requires v1.3.0
[79098fc4] Rmath v0.7.1
[f2b01f46] Roots v2.0.17
[7e49a35a] RuntimeGeneratedFunctions v0.5.12
[0bca4576] SciMLBase v1.94.0
[c0aeaf25] SciMLOperators v0.3.6
[30f210dd] ScientificTypesBase v3.0.0
[6c6a2e73] Scratch v1.2.0
[efcf1570] Setfield v1.1.1
[992d4aef] Showoff v1.0.3
[777ac1f9] SimpleBufferStream v1.1.0
[ce78b400] SimpleUnPack v1.1.0
[a2af1166] SortingAlgorithms v1.1.1
[276daf66] SpecialFunctions v2.3.0
[171d559e] SplittablesBase v0.1.15
[90137ffa] StaticArrays v1.6.2
[1e83bf80] StaticArraysCore v1.4.2
[64bff920] StatisticalTraits v3.2.0
[82ae8749] StatsAPI v1.6.0
[2913bbd2] StatsBase v0.34.0
[4c63d2b9] StatsFuns v1.3.0
[892a3eda] StringManipulation v0.3.0
[09ab397b] StructArrays v0.6.15
[2efcf032] SymbolicIndexingInterface v0.2.2
[3783bdb8] TableTraits v1.0.1
[bd369af6] Tables v1.10.1
[62fd8b95] TensorCore v0.1.1
[5d786b92] TerminalLoggers v0.1.7
[9f7883ad] Tracker v0.2.26
[3bb67fe8] TranscodingStreams v0.9.13
[28d57a85] Transducers v0.4.78
[410a4b4d] Tricks v0.1.7
[781d530d] TruncatedStacktraces v1.4.0
[fce5fe82] Turing v0.28.1
[5c2747f8] URIs v1.5.0
[1cfade01] UnicodeFun v0.4.1
[1986cc42] Unitful v1.16.2
[45397f5d] UnitfulLatexify v1.6.3
[013be700] UnsafeAtomics v0.2.1
[d80eeb9a] UnsafeAtomicsLLVM v0.1.3
[41fe7b60] Unzip v0.2.0
[efce3f68] WoodburyMatrices v0.5.5
[700de1a5] ZygoteRules v0.2.3
[6e34b625] Bzip2_jll v1.0.8+0
[83423d85] Cairo_jll v1.16.1+1
[2e619515] Expat_jll v2.5.0+0
⌃ [b22a6f82] FFMPEG_jll v4.4.2+2
[f5851436] FFTW_jll v3.3.10+0
[a3f928ae] Fontconfig_jll v2.13.93+0
[d7e528f0] FreeType2_jll v2.13.1+0
[559328eb] FriBidi_jll v1.0.10+0
[0656b61e] GLFW_jll v3.3.8+0
[d2c73de3] GR_jll v0.72.9+1
[78b55507] Gettext_jll v0.21.0+0
[7746bdde] Glib_jll v2.74.0+2
[3b182d85] Graphite2_jll v1.3.14+0
[2e76f6c2] HarfBuzz_jll v2.8.1+1
[1d5cc7b8] IntelOpenMP_jll v2023.2.0+0
[aacddb02] JpegTurbo_jll v2.1.91+0
[c1c5ebd0] LAME_jll v3.100.1+0
[88015f11] LERC_jll v3.0.0+1
[dad2f222] LLVMExtra_jll v0.0.23+0
[1d63c593] LLVMOpenMP_jll v15.0.4+0
[dd4b983a] LZO_jll v2.10.1+0
⌅ [e9f186c6] Libffi_jll v3.2.2+1
[d4300ac3] Libgcrypt_jll v1.8.7+0
[7e76a0d4] Libglvnd_jll v1.6.0+0
[7add5ba3] Libgpg_error_jll v1.42.0+0
[94ce4f54] Libiconv_jll v1.16.1+2
[4b2f31a3] Libmount_jll v2.35.0+0
[89763e89] Libtiff_jll v4.5.1+1
[38a345b3] Libuuid_jll v2.36.0+0
[856f044c] MKL_jll v2023.2.0+0
[e7412a2a] Ogg_jll v1.3.5+1
⌅ [458c3c95] OpenSSL_jll v1.1.22+0
[efe28fd5] OpenSpecFun_jll v0.5.5+0
[91d4177d] Opus_jll v1.3.2+0
[30392449] Pixman_jll v0.42.2+0
[c0090381] Qt6Base_jll v6.4.2+3
[f50d1b31] Rmath_jll v0.4.0+0
[a2964d1f] Wayland_jll v1.21.0+0
[2381bf8a] Wayland_protocols_jll v1.25.0+0
[02c8fc9c] XML2_jll v2.10.3+0
[aed1982a] XSLT_jll v1.1.34+0
[ffd25f8a] XZ_jll v5.4.4+0
[4f6342f7] Xorg_libX11_jll v1.8.6+0
[0c0b7dd1] Xorg_libXau_jll v1.0.11+0
[935fb764] Xorg_libXcursor_jll v1.2.0+4
[a3789734] Xorg_libXdmcp_jll v1.1.4+0
[1082639a] Xorg_libXext_jll v1.3.4+4
[d091e8ba] Xorg_libXfixes_jll v5.0.3+4
[a51aa0fd] Xorg_libXi_jll v1.7.10+4
[d1454406] Xorg_libXinerama_jll v1.1.4+4
[ec84b674] Xorg_libXrandr_jll v1.5.2+4
[ea2f1a96] Xorg_libXrender_jll v0.9.10+4
[14d82f49] Xorg_libpthread_stubs_jll v0.1.1+0
[c7cfdc94] Xorg_libxcb_jll v1.15.0+0
[cc61e674] Xorg_libxkbfile_jll v1.1.2+0
[12413925] Xorg_xcb_util_image_jll v0.4.0+1
[2def613f] Xorg_xcb_util_jll v0.4.0+1
[975044d2] Xorg_xcb_util_keysyms_jll v0.4.0+1
[0d47668e] Xorg_xcb_util_renderutil_jll v0.3.9+1
[c22f9ab0] Xorg_xcb_util_wm_jll v0.4.1+1
[35661453] Xorg_xkbcomp_jll v1.4.6+0
[33bec58e] Xorg_xkeyboard_config_jll v2.39.0+0
[c5fb5394] Xorg_xtrans_jll v1.5.0+0
[3161d3a3] Zstd_jll v1.5.5+0
⌅ [214eeab7] fzf_jll v0.29.0+0
[a4ae2306] libaom_jll v3.4.0+0
[0ac62f75] libass_jll v0.15.1+0
[f638f0a6] libfdk_aac_jll v2.0.2+0
[b53b4c65] libpng_jll v1.6.38+0
[f27f6e37] libvorbis_jll v1.3.7+1
[1270edf5] x264_jll v2021.5.5+0
[dfaa095f] x265_jll v3.5.0+0
[d8fb68d0] xkbcommon_jll v1.4.1+0
[0dad84c5] ArgTools v1.1.1
[56f22d72] Artifacts
[2a0f44e3] Base64
[ade2ca70] Dates
[8ba89e20] Distributed
[f43a241f] Downloads v1.6.0
[7b1f6079] FileWatching
[9fa8497b] Future
[b77e0a4c] InteractiveUtils
[4af54fe1] LazyArtifacts
[b27032c2] LibCURL v0.6.3
[76f85450] LibGit2
[8f399da3] Libdl
[37e2e46d] LinearAlgebra
[56ddb016] Logging
[d6f4376e] Markdown
[a63ad114] Mmap
[ca575930] NetworkOptions v1.2.0
[44cfe95a] Pkg v1.9.2
[de0858da] Printf
[3fa0cd96] REPL
[9a3f8284] Random
[ea8e919c] SHA v0.7.0
[9e88b42a] Serialization
[1a1011a3] SharedArrays
[6462fe0b] Sockets
[2f01184e] SparseArrays
[10745b16] Statistics v1.9.0
[4607b0f0] SuiteSparse
[fa267f1f] TOML v1.0.3
[a4e569a6] Tar v1.10.0
[8dfed614] Test
[cf7118a7] UUIDs
[4ec0a83e] Unicode
[e66e0078] CompilerSupportLibraries_jll v1.0.5+0
[deac9b47] LibCURL_jll v7.84.0+0
[29816b5a] LibSSH2_jll v1.10.2+0
[c8ffd9c3] MbedTLS_jll v2.28.2+0
[14a3606d] MozillaCACerts_jll v2022.10.11
[4536629a] OpenBLAS_jll v0.3.21+4
[05823500] OpenLibm_jll v0.8.1+0
[efcefdf7] PCRE2_jll v10.42.0+0
[bea87d4a] SuiteSparse_jll v5.10.1+6
[83775a58] Zlib_jll v1.2.13+0
[8e850b90] libblastrampoline_jll v5.8.0+0
[8e850ede] nghttp2_jll v1.48.0+0
[3f19e933] p7zip_jll v17.4.0+0
Info Packages marked with ⌃ and ⌅ have new versions available, but those with ⌅ are restricted by compatibility constraints from upgrading. To see why use `status --outdated -m`